19/04/2023
Print PageMultiplexed Ion Beam Imaging (MIBI) & its applications in cancer research
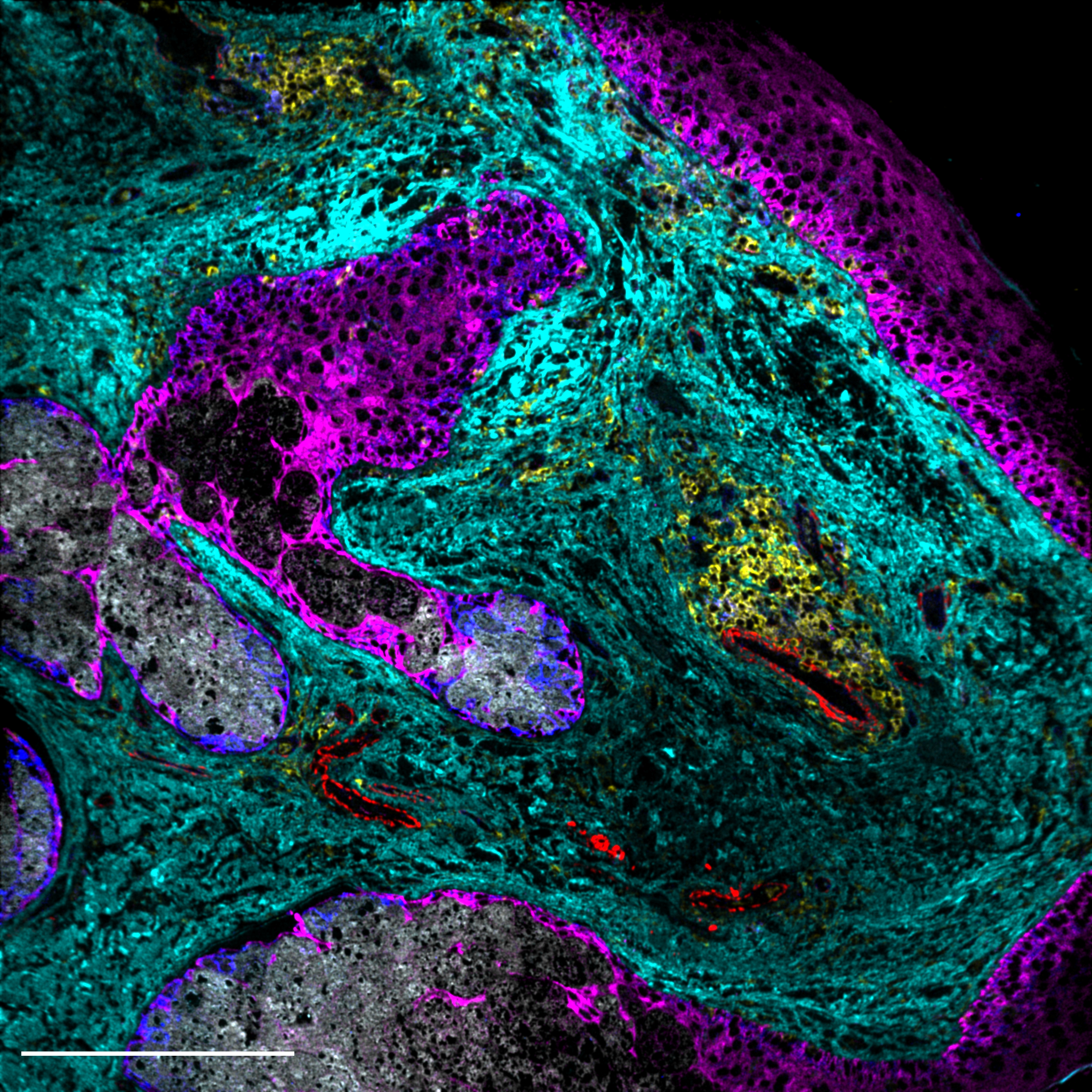
In order to answer questions about how immune cells interact with their local environment and how these interactions affect clinical outcomes in human cancers, Felix Hartmann uses two technologies to analyze immense data sets: Single Cell Mass Cytometry (CyTOF) and Multiplexed Ion Beam Imaging (MIBI). In his Tech Talk for the DKTK School of Oncology fellows he focuses on MIBI, which is a novel technology that enables high-dimensional proteomic imaging of standard tissue sections. In MIBI, antibodies conjugated to metal isotopes are quantified via time-of-flight mass spectrometry to visualize the spatial distribution of up to 40 proteins in parallel.
Since last year, this impressive imaging technology is feasible in Heidelberg in Felix Hartmann's lab –the only one of its kind in Europe. He uses MIBI frequently because it provides a unique combination of features:
- analysis of the spatial distribution of cells in tissues to identify tissue structure, multicellular communities, and cellular interactions
- quantification of proteins and posttranslational modifications
- adjustable resolution down to sub-cellular level
- no autofluorescence background
- the dynamic range spans five orders of magnitude
- tissue staining imaging of all targets is performed in a single step, and compatibility with a wide range of samples, including fixed-frozen and formalin-fixed paraffin-embedded (FFPE) tissue
Taken together, these features allow robust, sub-cellular imaging of cellular metabolism and phenotype in large sample collections and human clinical cohorts.
Scientists can select antibodies for targets and structures that are relevant to the individual question being addressed. One important question is how to analyze cellular metabolism, which has an impact on cancer. To answer this one approach by Felix Hartmann is to look at everything that regulates the metabolism, such as enzymes, signaling molecules, or transcription factors, and thus analyze the "metabolic regulome" at the single cell level. One finding is that T cell regulation is relevant to metabolism and that metabolic enzyme activity correlates with T cell activation. Metabolic analysis of the tumor microenvironment indicates the heterogeneity of T cell metabolic states: low expression of regulatory factors indicates a resting state of these cells. Thus, T cell metabolism reflects important functional states. Tissue visualization reveals spatially separated metabolic niches, e.g., differences between immune cells that are close to the tumor and those that are further away.
The group of Felix Hartmann mainly works with tissue samples of solid tumors and cooperates with clinicians and data scientists. At the end of his presentation, Felix Hartmann shared insights into current projects of individual members of his working group. One project focuses on mapping and quantifying metabolic states of immune cells across four human solid cancers. Another project deals with the identification of metabolic states that may be helpful in predicting therapeutic success and clinical outcome of patients. Finally, a third project addresses the analysis of metabolic niche formation in patient-derived tumor organoids by multiomic imaging.
About the Speaker: Dr. Felix Hartmann (DKTK Young Investigator Group - Systems Immunology and Single Cell Biology) received a B.Sc. and M.Sc. in Molecular Biotechnology from the University of Heidelberg, Germany, and his Ph.D. from the University of Zurich, Switzerland for his research on T cell effector functions in human autoimmune diseases. In 2017, he joined Stanford University as a postdoctoral fellow to study anti-tumor immunity and its metabolic regulation. Since 2021, Dr. Hartmann heads an independent DKTK Young Investigator Group at the DKFZ in Heidelberg. His research combines single-cell and imaging proteomic technologies with novel biological assays to reveal interactions of immune cells with their local environment and how these interactions impact clinical outcomes in human cancer. Most recently, he has developed a novel approach that enables analysis of cellular metabolism in individual cells and with spatial resolution.